Making a glass brain
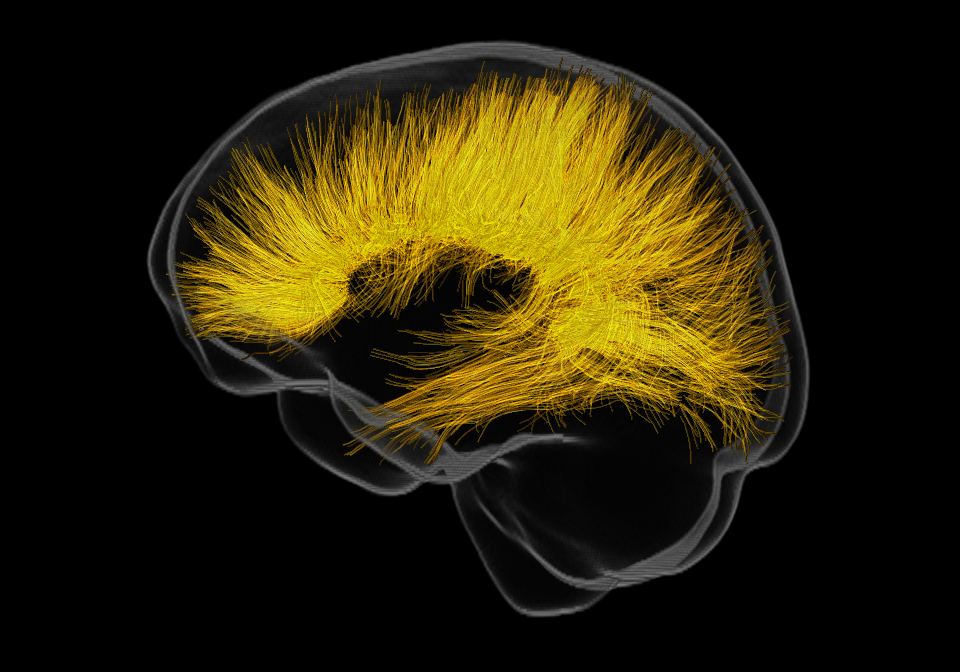
Everything looks better with glass
How can we give the deserved credit to white matter pathways in diffusion MRI? Easy: let’s throw away everything besides white matter. Jokes aside, tractography can already be a lot to digest, and sometimes we just need a rough reference to understand what we are looking at. If it is true that less is more, glass brains are a simple yet effective idea. In this post, I want to explore one way to make a smooth brain proposed by Thijs Dhollander: using the average from several brain masks, and then playing around with image operations to obtain a smooth and thin silhouette. A good example of this approach is showed in this paper by Remika Mito and colleagues.
The overall bash script to generate the glass brain and the related requirements are on the NeuroSnippets repository.
First steps
As sample data, I decided to use the dataset shared by Mario Mascalchi and colleagues on OpenNeuro, relying only on the first sessions of the healthy controls. This dataset can be easily downloaded through DataLad:
# Retrieving the data using DataLad
datalad install https://github.com/OpenNeuroDatasets/ds001378
cd ds001378
datalad get -J 4 sub-control*/ses-01/dwi/*
Once the data have been downloaded, I used MRtrix3 to:
- convert the data in MIF format, in order to embed the gradient information in the data;
- do a basic pre-processing (PCA denoising, Gibbs unringing, eddy current correction);
- compute an average volume from the b0 volumes;
- estimate a brain mask. All these steps are done in the following loop:
# Pre-processing using MRtrix3
for sub in $(ls -d sub-control*); do
mrconvert -fslgrad $sub/ses-01/dwi/${sub}_ses-01_dwi.bvec \
$sub/ses-01/dwi/${sub}_ses-01_dwi.bval $sub/ses-01/dwi/${sub}_ses-01_dwi.nii.gz \
$sub/ses-01/dwi/${sub}_ses-01_dwi.mif
dwidenoise $sub/ses-01/dwi/${sub}_ses-01_dwi.mif \
$sub/ses-01/dwi/${sub}_ses-01_dwi_desc-denoised.mif
mrdegibbs $sub/ses-01/dwi/${sub}_ses-01_dwi_desc-denoised.mif \
$sub/ses-01/dwi/${sub}_ses-01_dwi_desc-unringed.mif
dwipreproc $sub/ses-01/dwi/${sub}_ses-01_dwi_desc-unringed.mif \
$sub/ses-01/dwi/${sub}_ses-01_dwi_desc-preproc.mif -rpe_none -pe_dir PA
dwiextract -bzero $sub/ses-01/dwi/${sub}_ses-01_dwi_desc-preproc.mif - | \
mrmath - mean $sub/ses-01/dwi/${sub}_ses-01_dwi_desc-meanb0.mif -axis 3
dwi2mask $sub/ses-01/dwi/${sub}_ses-01_dwi_desc-preproc.mif \
$sub/ses-01/dwi/${sub}_ses-01_dwi_desc-mask.mif
done
Since I wanted a smooth result, just averaging volumes from different subjects more or less in the same location would not be great. One elegant way to align them using MRtrix3 is building a template from all the subjects and then align all the data to the template:
# Building a template with MRtrix3
mkdir -p ../template/meanb0s
mkdir ../template/masks
foreach sub-control* : ln -sr IN/ses-01/dwi/NAME_ses-01_dwi_desc-meanb0.mif \
../template/meanb0s/NAME.mif
foreach sub-control* : ln -sr IN/ses-01/dwi/NAME_ses-01_dwi_desc-mask.mif \
../template/masks/NAME.mif
population_template ../template/meanb0s -mask_dir ../template/masks \
../template/meanb0_template.mif
foreach sub-control* : mrregister IN/ses-01/dwi/NAME_ses-01_dwi_desc-meanb0.mif \
-mask1 IN/ses-01/dwi/NAME_ses-01_dwi_desc-mask.mif ../template/meanb0_template.mif \
-nl_warp IN/ses-01/dwi/NAME_ses-01_dwi_desc-warp_sub2tmp.mif \
IN/ses-01/dwi/NAME_ses-01_dwi_desc-warp_tmp2sub.mif
mkdir -p ../template/masks_tmp
foreach sub-control* : mrtransform IN/ses-01/dwi/NAME_ses-01_dwi_desc-mask.mif \
-warp IN/ses-01/dwi/NAME_ses-01_dwi_desc-warp_sub2tmp.mif -inter nearest \
-datatype bit ../template/masks_tmp/IN.mif
cd ../template
As the goal here is just in terms of visualization, for the sake of execution time I built the template out of the average b0 volumes. The population_template tool actually allows to do so much more, as shown in this tutorial.
Where is the glass?
Now that we have all the masks, it is finally time to make things even smoother! This is what we will do:
- average all the masks aligned with the template;
- upsample the mean mask;
- use a Gaussian filter to smooth the upsampled mask;
- use a threshold to obtain a neat black-and-white result;
- dilate the previous volume;
- subtract the volume obtained in point 4 from the dilated volume.
# Making a glass brain!
mrmath masks_tmp/* mean mask_mean.mif
mrresize mask_mean.mif -scale 5 mask_up.mif
mrfilter mask_up.mif smooth -stdev 2 mask_smooth.mif
mrthreshold mask_smooth.mif -abs 0.5 mask_thres.mif
maskfilter mask_thres.mif dilate -npass 2 mask_dilated.mif
mrcalc mask_dilated.mif mask_thres.mif -subtract glass_brain.mif
And we have our smooth brain surface! Playing with the alpha control in mrview will allow us to make it glassy. Now that we have a glass brain, we need something to put inside it!
Filling (!) the glass brain
Let’s reconstruct a corpus callosum to be displayed in our glass brain. I selected the first subject as an example, estimated the response function using the Tournier algorithm, and then estimated the fiber orientration distribution. To quickly reconstruct the corpus callosum without having to define regions of interest or anything like that, I used TractSeg. The very final step is to align the reconstructed corpus callosum with the template and therefore the glass brain.
# Reconstructing the Corpus Callosum with TractSeg
cd ../ds001378/sub-control01/ses-01/dwi
dwi2response tournier sub-control01_ses-01_dwi_desc-preproc.mif \
sub-control01_ses-01_dwi_desc-response.txt
dwiextract sub-control01_ses-01_dwi_desc-preproc.mif - | dwi2fod msmt_csd - \
sub-control01_ses-01_dwi_desc-response.txt \
sub-control01_ses-01_dwi_desc-wmfod.mif \
-mask sub-control01_ses-01_dwi_desc-mask.mif
sh2peaks sub-control01_ses-01_dwi_desc-wmfod.mif \
sub-control01_ses-01_dwi_desc-peaks.nii.gz
TractSeg -i sub-control01_ses-01_dwi_desc-peaks.nii.gz
TractSeg -i sub-control01_ses-01_dwi_desc-peaks.nii.gz --output_type TOM
tckgen -algorithm FACT -angle 20 -seed_image sub-control01_ses-01_dwi_desc-mask.mif \
tractseg_output/TOM/CC.nii.gz sub-control01_ses-01_dwi_desc-CC.tck
tcktransform sub-control01_ses-01_dwi_desc-CC.tck \
sub-control01_ses-01_dwi_desc-warp_tmp2sub.mif ../../../../template/CC.tck
Now you have a corpus callosum to put inside your glass brain!
Useful references
- “Diffusion MRI visualization”
- SCA2 Diffusion Tensor Imaging dataset
- “Histogram analysis of DTI-derived indices reveals pontocerebellar degeneration and its progression in SCA2”
- “Fibre-specific white matter reductions in Alzheimer’s disease and mild cognitive impairment”
- A more complex approach to template-based analysis of dMRI data
