Histogrammers: a dashboard to quickly check MRI data
All it takes is an ROI
When one is dealing with MRI data (especially in the case of quantitative MRI), it is usually a good idea to check how the data are distributed. An example: you picked a myelin water fraction (MWF) map - it is reasonable to expect that the values will be higher in white matter than in grey matter. After this very first quality check, probably you may want to check how things change in different white matter structures. An easy way to do that would be plotting a histogram of the data distribution in a given region - except that then you need to fire up (*gasp*) MATLAB. You can plot some histograms in most neuroimaging packages (first tools that come to mind are fsleyes, freeview, itksnap), but unfortunately it is not immediate to create a histogram of a specific area. Truth to be told, you also need to think about defining the ROIs, or picking an atlas. It started as a quick check, but suddenly you are in the middle of processing the whole thing. Is there an easier alternative?
In this post I show how to leverage the amazing Dash framework to put together a tailored dashboard in more or less 100 lines of code - it will allow us to just sketch on top of the image to define our region of interest. To render the dashboard, we can use a Jupyter Notebook (as I will do here, and as you can test immediately on MyBinder or Colab) or run it as a flask-based web app. Both these examples and the related requirements are on the NeuroSnippets repository.
The first blow is half the dashboard
For this example, I decided to use the quantitative T1 map shared by José P. Marques on the MP2RAGE script repository - MP2RAGE is both a common and simple way to acquire T1-weighted data that can later be used to estimate a map of the longitudinal relaxation time. For pure convenience, I cropped and reoriented the original T1 map - the steps are included in a very brief script on GitHub.
So let’s start writing down the modules we need and some utility functions:
from jupyter_dash import JupyterDash
import dash
import dash_core_components as dcc
import dash_html_components as html
import nibabel as nib
import numpy as np
import plotly.express as px
from dash.dependencies import Input, Output, State
from scipy import ndimage
from skimage import draw
def path_to_indices(path):
"""From SVG path to numpy array of coordinates, each row being a (row, col) point
"""
indices_str = [
el.replace('M', '').replace('Z', '').split(',') for el in path.split('L')
]
return np.floor(np.array(indices_str, dtype=float)).astype(np.int)
def path_to_mask(path, shape):
"""From SVG path to a boolean array where all pixels enclosed by the path
are True, and the other pixels are False.
"""
cols, rows = path_to_indices(path).T
rr, cc = draw.polygon(rows, cols)
mask = np.zeros(shape, dtype=np.bool)
mask[rr, cc] = True
mask = ndimage.binary_fill_holes(mask)
return mask
These two functions come directly from the Dash documentation, and they will be very useful to first convert any SVG path drawn on top of the image to a list of spatial coordinates, and then use these coordinates to make a mask of our hand-drawn ROI.
The next step is loading the data and start figuring out the main elements of this dashboard:
# Loading the data and creating the figures
img = nib.load('T1map_cropped_reoriented.nii.gz')
data = img.dataobj
default_view = 2
default_slice = int(data.shape[default_view]/2)
view = np.take(data, default_slice, axis=default_view)
view = view.T
fig = px.imshow(view, binary_string=True, origin='lower')
fig.update_layout(dragmode='drawopenpath',
newshape=dict(opacity=0.8, line=dict(color='red', width=2)))
config = {
'modeBarButtonsToAdd': [
'drawopenpath',
'drawclosedpath',
'eraseshape'
]
}
fig_hist = px.histogram(data.get_unscaled().ravel())
fig_hist.update_layout(showlegend=False)
app = JupyterDash(__name__)
server = app.server
Once the 3D volume is loaded using nibabel, we define preliminarly which plane of view (in this case, the axial one) and which slice (one in the middle) to visualize - we will deal in a bit on how to change those. Then, we first create a figure out of this slice with plotly_express.imshow(), adding the elements we will need to draw on top of it, and then we create a histogram, that at the beginning will show the T1 distribution of the overall volume. And finally, we create the app object.
It is time to define the layout of our dashboard:
# Defining the layout of the dashboard
app.layout = html.Div([html.Div([
html.Div(
[dcc.Dropdown(id="plane-dropdown", options=[
{'label': 'Axial', 'value': 2},
{'label': 'Coronal', 'value': 1},
{'label': 'Sagittal', 'value': 0}],
value=2),
dcc.Graph(id='graph-mri', figure=fig, config=config),
dcc.Slider(
id='slice-slider',
min=0,
max=data.shape[default_view] - 1,
value=int(data.shape[default_view]/2),
step=1)],
style={'width': '60%', 'display': 'inline-block', 'padding': '0 0'},
),
html.Div(
[dcc.Graph(id='graph-histogram', figure=fig_hist)],
style={'width': '40%', 'display': 'inline-block', 'padding': '0 0'},
), html.Div(id='test')
])
])
The general structure is based on two html.Div components (which wrap the related div elements in HTML), the first one containing a dropdown menu (that we will use to change the plane of view), a graph embedding our MRI slice, and a slider (that we will use to move across the slices); in the second element, we have a graph embedding the histogram. Using the style parameters, we arrange them in order to have the first component on left and the second one on the right. Well, the dashboard is designed!
“And yet it does not move”
At the moment, everything is quite static: if we try to run the code so far, and make the app rendering, we will be able to see the middle axial slice as already defined, and the histogram for the overall volume. We can already draw on top of the MRI slice, but nothing happens. It’s time to make things interactive!
To be able to navigate the MRI volume and to make our drawings triggering an update in the histogram, we need to define two callback functions - they are identified as such because of the @app.callback decorator. Let’s have a look at the first one:
@app.callback(
[Output('graph-mri', 'figure'),
Output('slice-slider', 'max'),
Output('slice-slider', 'value')],
[Input('plane-dropdown', 'value'),
Input('slice-slider', 'value')],
prevent_initial_call=True)
def update_plane(plane, vol_slice):
ctx = dash.callback_context
if ctx.triggered[0]['prop_id'].split('.')[0]=='plane-dropdown':
vol_slice = int(data.shape[plane]/2)
view = np.take(data, vol_slice, axis=plane)
view = view.T
fig = px.imshow(view, binary_string=True, origin='lower')
fig.update_layout(dragmode='drawopenpath',
newshape=dict(opacity=0.8, line=dict(color='red', width=2)))
return [fig, data.shape[plane] - 1, vol_slice]
This function handles the input from two components (the dropdown menu and the slider) and uses as outputs three properties (the graph content, the slider maximum value, and the slider current value). Why is that? As a general case, the size of the volume in each dimension will not be the same, so the slider needs to be updated accordingly. Inside the function, before dealing with updating the figure, we need also to decide if the current position is still acceptable. Consider this case: if the slider is towards the very end in a large dimension, switching the plane of view to a smaller dimension will cause some troubles. To avoid this, we use the callback_context to check which input component caused the triggering of the function, and if it is indeed the dropdown menu, we just reposition the slider in the middle accordingly to the new plane selected. As we now have a nice way to navigate the MRI volume, we are just missing the connection with the histogram:
@app.callback(
Output('graph-histogram', 'figure'),
Input('graph-mri', 'relayoutData'),
[State('plane-dropdown', 'value'),
State('slice-slider', 'value')],
prevent_initial_call=True)
def histo_from_annotation(relayout_data, current_plane, current_slice):
if 'shapes' in relayout_data:
last_shape = relayout_data['shapes'][-1]
view = np.take(data, current_slice, axis=current_plane)
view = view.T
mask = path_to_mask(last_shape['path'], view.shape)
fig = px.histogram(view[mask])
fig.update_layout(showlegend=False)
return fig
else:
return dash.no_update
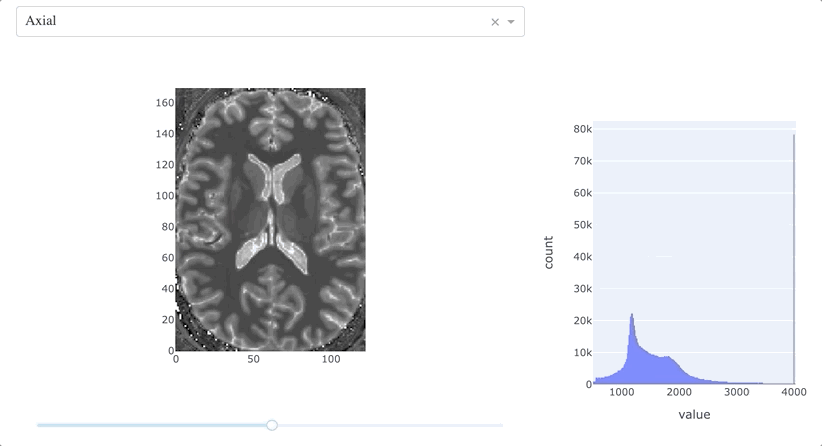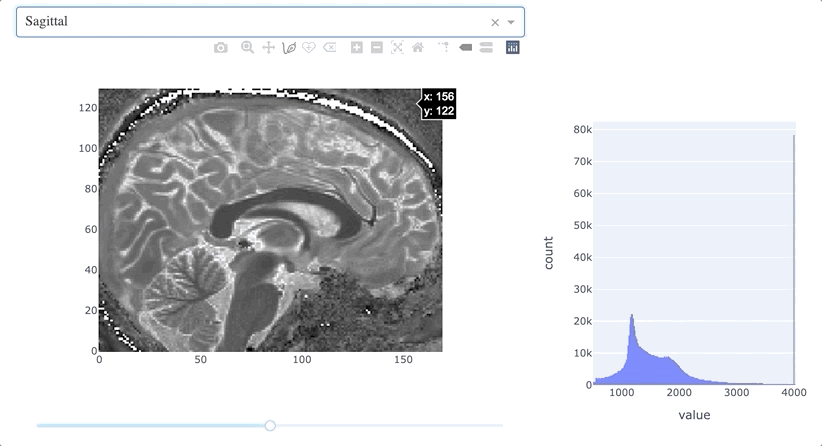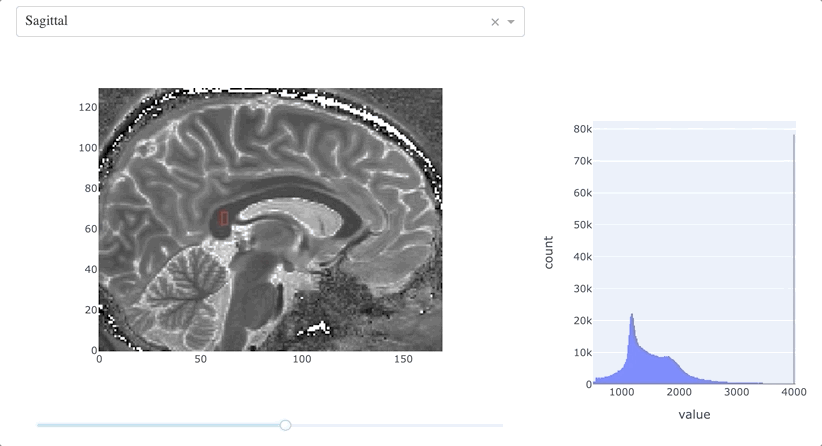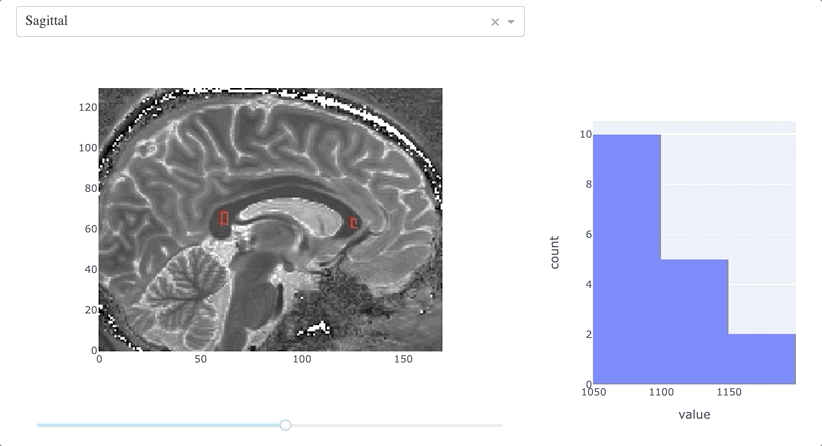
This function couples the MRI slice and the histogram:
- retrieves the most recent path drawn on the slice;
- computes a mask using
path_to_mask(); - generates a histogram of the values within the mask.
The decorator in this case does not only defines input and output (respectively the MRI slice and the histogram), but also takes into account the state of two objects, the dropdown and slider: this is to make sure that we always refer to the current plane and slice when we create and apply the mask.
Everything is ready. It is now just a matter of rendering our dashboard:
app.run_server(mode="inline")
